Playing Dominos: How an Artificial Protein Emerges from Fitting Together Individual Components
Heidelberg scientists develop AI model for producing new molecular machines in a targeted way
The targeted engineering of artificial proteins with unique properties – that is possible with the assistance of a novel method developed by a research team led by Prof. Dr Dominik Niopek at the Institute of Pharmacy and Molecular Biotechnology (IPMB) of Heidelberg University. It centers around a new AI model. This allows for forecasting how two proteins have to be fitted together at the molecular level from individual parts – subunits – in order to engineer a functional, adjustable new protein. Called ProDomino, this open-source tool opens up many different possible applications in biotechnology and medicine.
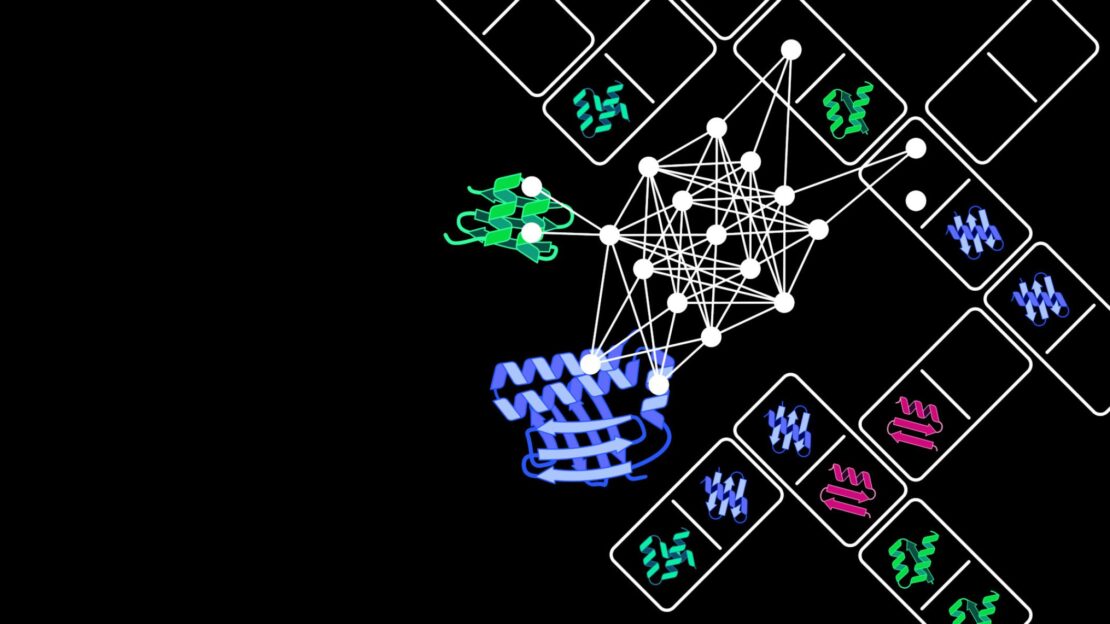
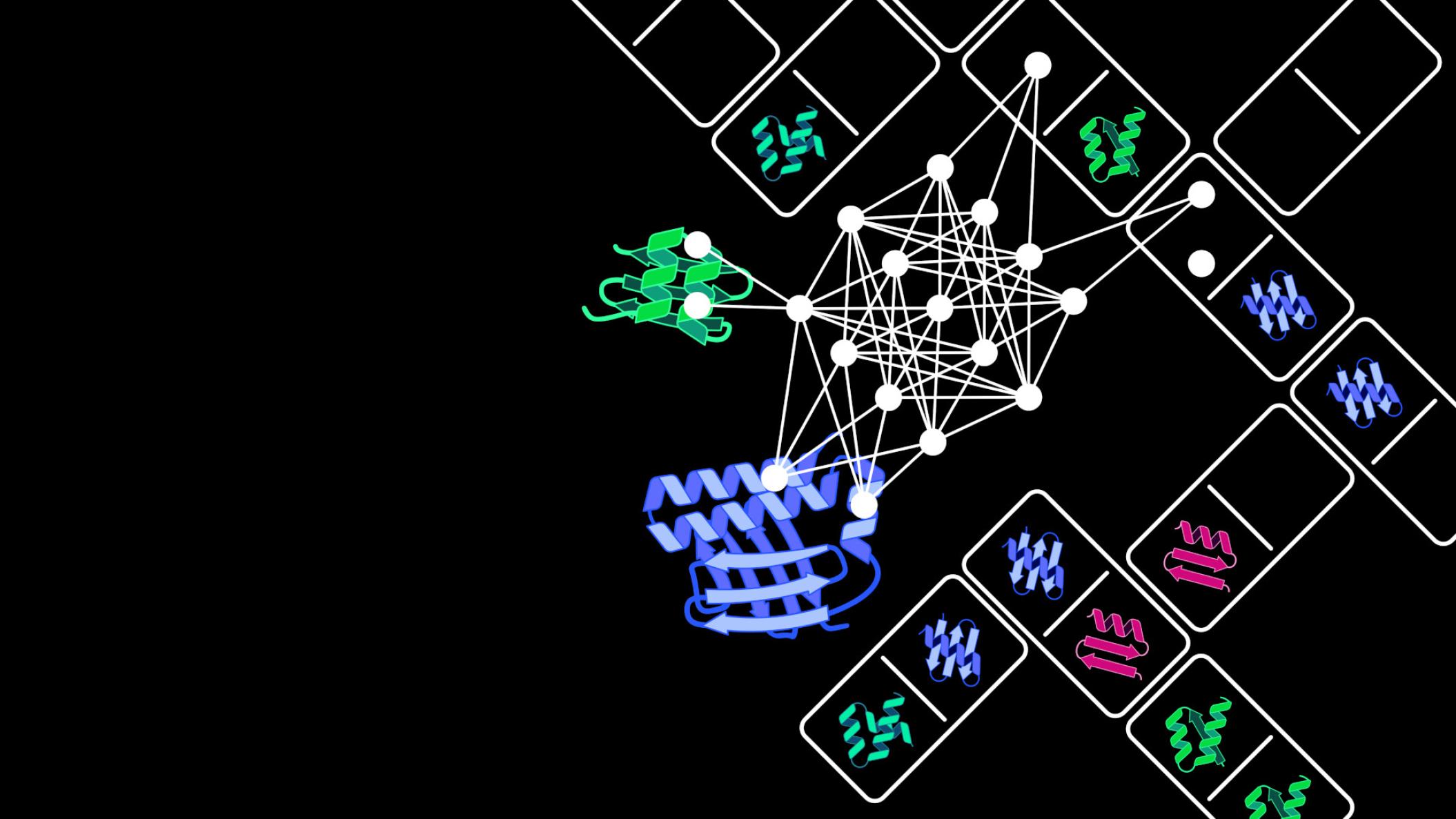
At the cell level, proteins drive numerous vital processes such as gaining energy or increasing genetic material and are hence frequently called the “molecular machines” of the cells. They consist of interlinked subunits known as domains. “Like components of a technical machine these domains have specific functions. For example, they enable the proteins to recognize external signals such as light or temperature, or to catalyze chemical reactions with the assistance of proteins,” underlines Prof. Niopek, who heads the Pharmaceutical Biology department at the IPMB. Novel proteins frequently arise naturally through existing domains connecting up in new combinations.
Inspired by these evolutionary innovation processes, Dominik Niopek and his team have developed an AI model on the artificial recombination of protein domains. Extensive and high-quality data sets are essential to develop and optimize these complex computer models. “Through the automated analysis of huge protein databases we were able to produce a tailored data set with over 100,000 proteins. It reflects the enormous variety of natural domain combinations and was the central foundation for training our AI model,” says Benedict Wolf, a doctoral candidate in Prof. Niopek’s team. The Protein Domain Insertion Optimizer, abbreviated as ProDomino, can forecast which protein domains – like domino tiles – have to be fitted together to engineer a merged protein with new properties.
With the aid of ProDomino, for example, protein sensors, which react to stimuli like light or chemical substances, can be combined with effector proteins to control cellular processes. Consequently, the Heidelberg scientists succeeded in coupling chemosensitive domains with the “genetic scissors” CRISPR-Cas, thereby generating artificial CRISPR-Cas variants. These hybrid proteins can be deliberately turned on or off and so could considerably increase the safety of modern genome editing procedures, explains Dr Jan Mathony, group leader in Prof. Niopek’s department. The researchers have successfully tested their AI model on different proteins and made the software available to scientists as an open-source tool.
“Our AI model allows for producing artificial proteins not only more easily but also in a more targeted way than before. This opens up new opportunities for improving the precision of protein-based applications in biotechnology and medicine, or possibly developing completely new therapy approaches,” says Prof. Niopek.
This research study was funded by the European Research Council (ERC) of the European Union. The results have been published in the journal “Nature Methods”.
Original Publication
B. Wolf, P. Shehu, L. Brenker, A.L. von Bachmann, A.S. Kroell, N. Southern, S. Holderbach, J. Eigenmann, S. Aschenbrenner, J. Mathony, D. Niopek: Rational engineering of allosteric protein switches by in silico prediction of domain insertion sites. Nature Methods (2025)




