RNA in action: filming a ribozyme’s self-assembly
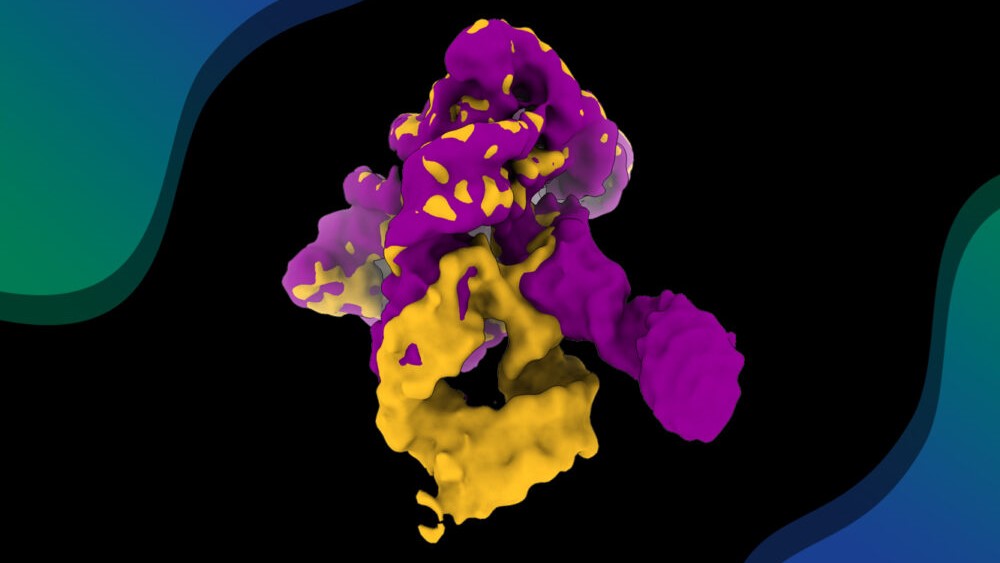
Researchers have visualized, in unprecedented detail, how a large RNA molecule assembles itself into a functional machine

RNA is a central biological molecule, now widely harnessed in medicine and nanotechnology. Like proteins, RNA often gets its function from its three-dimensional structure. A recent study in Nature Communications has captured, for the first time, a ribozyme in motion — almost frame by frame. The researchers recorded how this tiny RNA machine folds, flexes, and assembles itself, revealing its intricate choreography in unprecedented detail.
Using state-of-the-art techniques — cryo-electron microscopy (cryo-EM), small-angle They captured the dynamic ‘behind-the-scenes’ process by which the self-splicing ribozyme folds into its functional structure. The research was led by the team of Marco Marcia , former EMBL Group Leader and currently Associate Professor and SciLifeLab Group leader at Uppsala University , Sweden.
This breakthrough was made possible thanks to the cutting-edge facilities and expert services at EMBL Grenoble, which enabled the integration of advanced structural biology methods with RNA biochemistry and enzymology. The Marcia group also benefited from close collaboration with the Center for Structural Systems Biology (CSSB) Hamburg, where innovative cryo-EM image processing approaches tailored for this specific project were developed, and the Istituto Italiano di Tecnologia (IIT), which provided high-level molecular simulation expertise.
“Determining RNA structures is a challenging task – the inherent flexibility and negative charge make RNA a notoriously difficult target for structural studies,” said Shekhar Jadhav, former Predoctoral Fellow at EMBL Grenoble, now a postdoc at Uppsala University, Sweden. “Persistent efforts and extensive screening on electron microscopes ultimately led us to visualize elusive RNA dynamics.”
How one domain orchestrates the RNA storyline
At the heart of this production is Domain 1 (D1), the ribozyme’s central scaffold and, as it turns out, its director. This domain acts as a molecular gate, cueing the other domains (D2, D3, D4) to enter at precisely the right moment during the folding process.
Subtle movements in key parts of the D1 molecule prompt one of its sections to open up and make way for the next. Each domain joins the scene only when the previous one is correctly in place, creating a seamless sequence of molecular choreography that prevents structural errors and ensures a flawless finale: the formation of a structure that can catalyze a chemical reaction, essential to the ribozyme’s function.
Capturing the hidden takes
By analyzing hundreds of thousands of single RNA particles, the team reconstructed intermediate ‘takes’ that were invisible in static crystal structures. These fleeting frames show how the RNA explores alternative poses before settling into its final conformation.
“To capture these fleeting frames, we had to develop novel cryo-EM image-processing strategies,” said Maya Topf , Group Leader at CSSB , Professor at the University Medical Center Hamburg-Eppendorf, and a collaborator on the study. “This is a great example of how computational innovation and high-quality cryoEM data can reveal the hidden conformations of molecular machines.”
SAXS data and molecular dynamics simulations helped the scientists refine each frame and assemble the storyline.
“One major strength of this work is the synergy between these cutting-edge new structural data on RNA and our advanced molecular simulations of this challenging system,” said Marco De Vivo , Head of Molecular Modeling and Drug Discovery Lab and Associate Director for Computation of Institu Italiano di Technologia in Genoa, and one of the collaborators on this study. “This combined approach has clarified, at an unprecedented atomistic level of detail, the dynamic that drives the entire assembly of this RNA molecule, which now opens new avenues for drug discovery efforts targeting RNA.”
From ancient scripts to modern spin-offs
Group II introns, the ribozymes featured in this molecular film, are thought to be the ancestors of the spliceosome, the complex machinery that edits RNA in human cells. By revealing how these molecules fold efficiently and avoid kinetic traps, the study provides new insight into how early RNA-based life may have evolved its editing tools. Beyond evolutionary lore, this work also sets the stage for RNA design and engineering – guiding how future biotechnologies might script RNA molecules to fold correctly for use in therapeutics or nanobiotechnology.
Opening the door to RNA AI
The detailed datasets and molecular mechanisms discovered in this study offer a valuable benchmark for training and testing AI models. Some of the RNA structures resolved here have already been used in international CASP competitions — the same predictive challenge that gave rise to AlphaFold — as recently described in the journal Proteins .
“This work is expected to play a key role in shaping artificial intelligence approaches to RNA structure prediction, paving the way towards a new ‘AlphaFold for RNA’.” Marcia said.
This convergence of experimental precision and machine learning marks a new phase for RNA structural biology, where AI and cryo-EM can learn from each other to predict, visualize, and understand the dynamics of life’s most versatile molecules.




